Working with categorical data + Saving data
Overview
Teaching: 30 min
Exercises: 15 minQuestions
How to fix common typos in character variables?
How to reorder values in ordinal categorical variables?
How to save data into a file?
Objectives
Discuss some common issues with data cleaning and use functions from the
stringrpackage to help solve them.Use factors to order categories and encode ordinal data.
Save data frame into a file.
In this lesson we’re going to learn how to use the dplyr package to manipulate columns
of our data.
As usual when starting an analysis on a new script, let’s start by loading the packages and reading the data. In this lesson we’re going to use the full dataset with data from 1960 to 2010:
library(tidyverse)
# Read the data, specifying how missing values are encoded
gapminder1960to2010 <- read_csv("data/raw/gapminder1960to2010_socioeconomic.csv",
na = "")
Manipulating categorical data
The stringr package provides several functions to manipulate strings (i.e. character values).
The functions from that package start with the word str_, so they are easy to identify.
For example, let’s have a look at the distinct values of the income_groups variable:
gapminder1960to2010 %>%
distinct(income_groups)
# A tibble: 4 x 1
income_groups
<chr>
1 low_income
2 lower_middle_income
3 upper_middle_income
4 high_income
Let’s say we would instead want to have the values “low”, “lower middle”, “upper middle”
and “high”. With the help of some str_*() functions we can do this in two steps:
- remove the string “_income”
- replace the underscore “_” with a space “ “
gapminder1960to2010 %>%
# remove the word "_income" from the income_groups values; and then...
mutate(income_groups = str_remove(income_groups, "_income")) %>%
# replace the "_" with a hyphen
mutate(income_groups = str_replace(income_groups, "_", " ")) %>%
# check the distinct values for income_groups
distinct(income_groups)
# A tibble: 4 x 1
income_groups
<chr>
1 low
2 lower middle
3 upper middle
4 high
Exercise
Create a new table called
gapminder_clean, which fulfils the following requirements:
- Fix any typos in the
main_religionvalues. All values should be in Title Case. (hint:str_squish()andstr_to_title())- The
world_regioncolumn contains values with a space between words (not “_”) and in Title Case. (hint:str_to_title()andstr_replace_all())- The
income_groupscolumn contains the categories: “Low”, “Lower Middle”, “Upper Middle”, “High”. (hint:str_remove(),str_to_title()andstr_replace_all())If you haven’t done so already, also make sure that
life_expectancy_femaleis numeric (as.numeric()) and thatlife_expectancy_malehas values “-999” encoded asNA(ifelse()).The final table should contain 9843 observations (rows) and 13 variables (columns), 4 of them character, 1 logical and the rest numeric.
Answer
gapminder_clean <- gapminder1960to2010 %>% # fix typos in main_religion and world region mutate(main_religion = str_to_title(str_squish(main_religion)), world_region = str_to_title(str_replace_all(world_region, "_", " "))) %>% # fit typos in income groups, which needs more steps mutate(income_groups = str_remove(income_groups, "_income")) %>% mutate(income_groups = str_to_title(str_replace_all(income_groups, "_", " "))) %>% # fix/create numeric variables mutate(life_expectancy_female = as.numeric(life_expectancy_female), life_expectancy_male = ifelse(life_expectancy_male == -999, NA, life_expectancy_male))Finally, check that the final table contains the right number of rows, columns and variable types using
str():str(gapminder_clean)tibble [9,843 × 13] (S3: spec_tbl_df/tbl_df/tbl/data.frame) $ country : chr [1:9843] "Afghanistan" "Afghanistan" "Afghanistan" "Afghanistan" ... $ world_region : chr [1:9843] "South Asia" "South Asia" "South Asia" "South Asia" ... $ year : num [1:9843] 1960 1961 1962 1963 1964 ... $ children_per_woman : num [1:9843] 7.45 7.45 7.45 7.45 7.45 7.45 7.45 7.45 7.45 7.45 ... $ life_expectancy : num [1:9843] 39.3 40 40.8 41.5 42.2 ... $ income_per_person : num [1:9843] 2744 2702 2683 2665 2649 ... $ is_oecd : logi [1:9843] FALSE FALSE FALSE FALSE FALSE FALSE ... $ income_groups : chr [1:9843] "Low" "Low" "Low" "Low" ... $ population : num [1:9843] 8996967 9169406 9351442 9543200 9744772 ... $ main_religion : chr [1:9843] "Muslim" "Muslim" "Muslim" "Muslim" ... $ child_mortality : num [1:9843] 357 351 345 340 335 ... $ life_expectancy_female: num [1:9843] 33.3 33.8 34.4 34.9 35.4 ... $ life_expectancy_male : num [1:9843] 31.7 32.2 32.7 33.2 33.7 ... - attr(*, "spec")= .. cols( .. country = col_character(), .. world_region = col_character(), .. year = col_double(), .. children_per_woman = col_double(), .. life_expectancy = col_double(), .. income_per_person = col_double(), .. is_oecd = col_logical(), .. income_groups = col_character(), .. population = col_double(), .. main_religion = col_character(), .. child_mortality = col_double(), .. life_expectancy_female = col_character(), .. life_expectancy_male = col_double() .. )
Using factors to encode ordinal categorical data
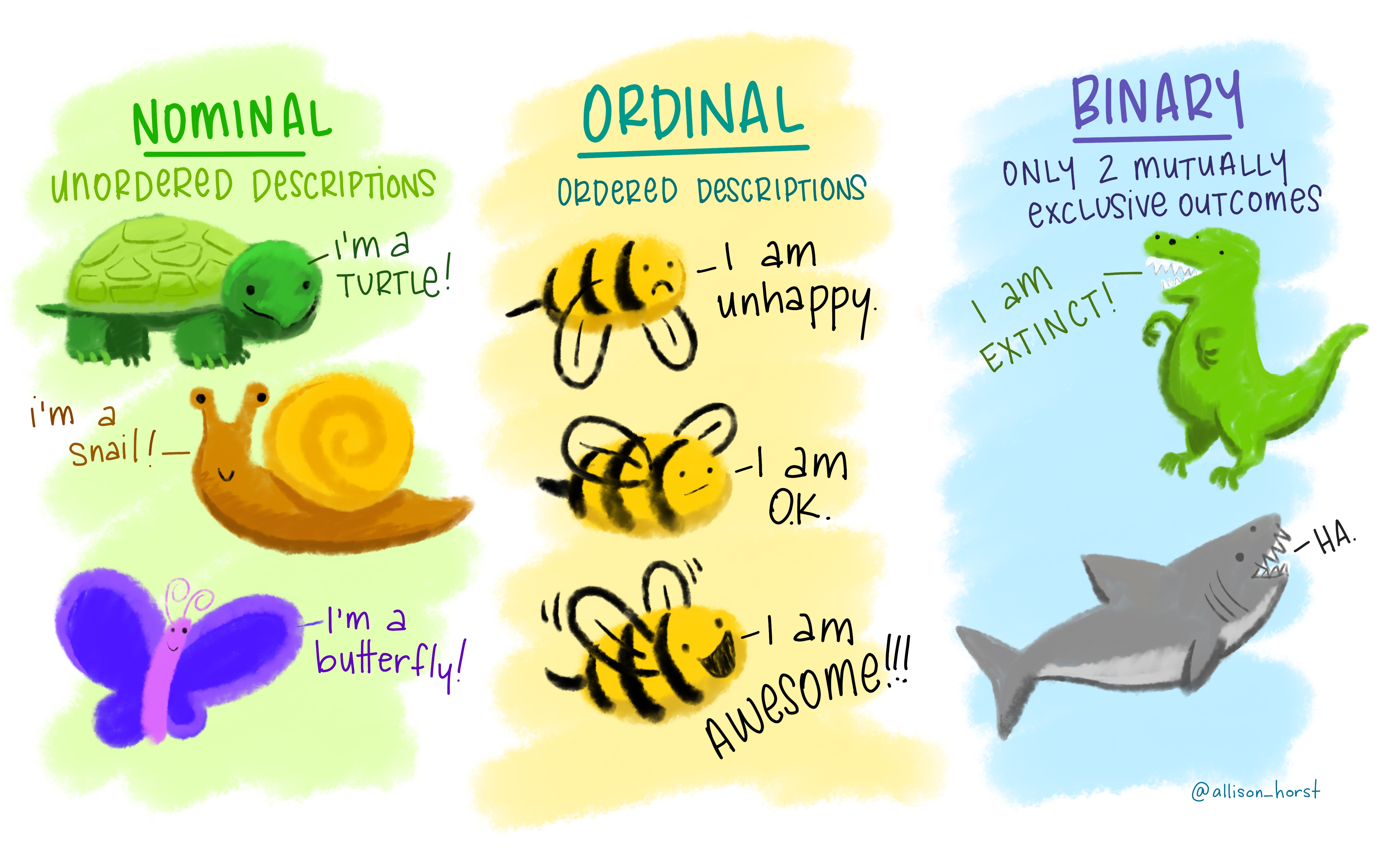
As we discussed in a previous episode, categorical data come in three flavours: nominal, ordinal and binary.
While character vectors can be used to encode non-ordered (nominal) categories,
they are not suitable to encode ordered ones. For this, we need to use factors, which
are a special type of vector that stores categorical data.
Here is an example using a character vector:
mood <- c("unhappy", "awesome", "ok", "awesome", "unhappy")
# convert mood character vector to a factor
factor(mood)
[1] unhappy awesome ok awesome unhappy
Levels: awesome ok unhappy
Once created, factors can only contain a pre-defined set of values, known as levels, which correspond to the unique values in the data. By default, R always sorts levels in alphabetical order, like in the example above.
Sometimes, the order of the levels does not matter, other times you might want to specify the order because it is meaningful (e.g., “low”, “medium”, “high”), it improves your visualization, or it is required by a particular type of analysis.
Here is how we would reorder the levels of the mood vector:
factor(mood, levels = c("unhappy", "ok", "awesome"))
[1] unhappy awesome ok awesome unhappy
Levels: unhappy ok awesome
The forcats package (part of tidyverse) provides several other functions to
manipulate factors. These functions all start with fct_, so they are easy to
identify. Look at the package documentation to
learn more about it.
Exercise
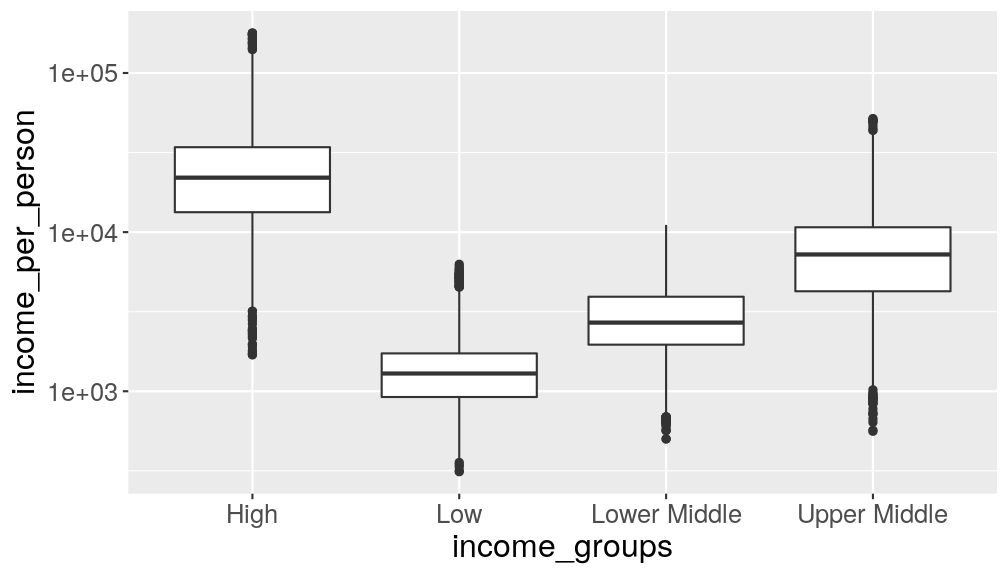
Take the following boxplot showing the distribution of income, per income groups:
gapminder_clean %>% ggplot(aes(income_groups, income_per_person)) + geom_boxplot() + scale_y_continuous(trans = "log10")
The ordering of the categories on the x-axis is alphabetical. In this case, it would make sense to change this order, to reflect it’s ranking.
Use the
factor()function withinmutate()to modify theincome_groupsvariable to have a more logical order.Answer
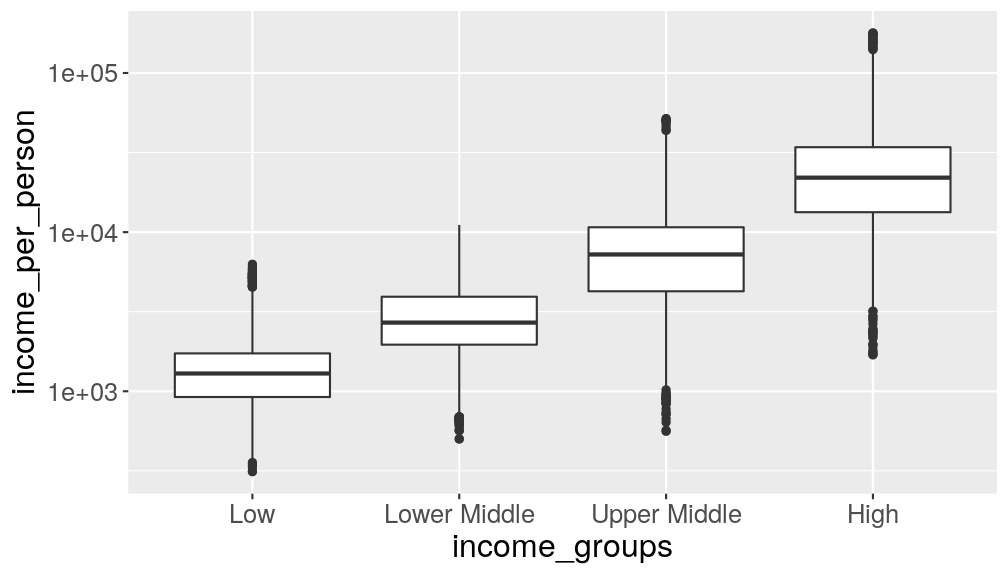
We can do this by mutating the variable into a factor, where we specify the levels manually:
gapminder_clean %>% # convert income groups to a factor mutate(income_groups = factor(income_groups, levels = c("Low", "Lower Middle", "Upper Middle", "High"))) %>% # make the graph ggplot(aes(income_groups, income_per_person)) + geom_boxplot() + scale_y_continuous(trans = "log10")
Saving data
Now that we have a clean version of our table, it’s a good idea to save it for
future use. You can use the write_*() family of functions to save data in a variety
of formats.
Let’s use write_csv() as an example.
The write_csv() function needs the name of the table you want to save and then
path to the file you want to save it in (don’t forget the file extension!):
write_csv(gapminder_clean, "data/processed/gapminder1960to2010_clean.csv")
There are many other functions for saving data, you can check the documentation
with ?write_delim.
Data Tip: Cleaning Data
The infamous 80/20 rule in data science suggests that about 80% of the time is spend preparing the data for analysis. While this is not really a scientific rule, it does have some relation to the real life experience of data analysts.
Although it’s a lot of effort, and usually not so much fun, if you make sure to clean and format your data correctly, it will make your downstream analysis much more fluid, fruitful and pleasant.
Key Points
Use functions from the
stringrpackage to manipulate strings. All these functions start withstr_, making them easy to identify.Use factors to encode ordinal variables, ensuring the levels are set in a logical order.