Manipulating observations (rows) with `dplyr`
Overview
Teaching: 40 min
Exercises: 25 minQuestions
How to order rows in a table?
How to retain only unique rows (no duplicates)?
How to identify observations of a dataset that fulfill certain conditions?
Objectives
Apply the functions
arrange()distinct()andfilter()to operate on rows.Understand how logical comparisons are made and the logical data type.
Remember and distinguish between different types of logical operators.
Use conditional operations to obtain specific observations from data using the
filter()function.Combine the
filter()andis.na()functions to remove missing values.Use the
ifelse()function to highlight specific observations.
In this lesson we’re going to learn how to use the dplyr package to manipulate rows
of our data.
As usual when starting an analysis on a new script, let’s start by loading the packages and reading the data. We will continue with gapminder data from 1960 to 2010:
library(tidyverse)
# Read the data, specifying how missing values are encoded
gapminder1960to2010 <- read_csv("data/raw/gapminder1960to2010_socioeconomic.csv",
na = "")
Ordering Rows
Or order the rows of a table based on certain variables, we can use the arrange()
function:
gapminder1960to2010 %>%
select(country, world_region, year) %>%
# order by year
arrange(year)
# A tibble: 9,843 x 3
country world_region year
<chr> <chr> <dbl>
1 Afghanistan south_asia 1960
2 Angola sub_saharan_africa 1960
3 Albania europe_central_asia 1960
4 Andorra europe_central_asia 1960
5 United Arab Emirates middle_east_north_africa 1960
6 Argentina america 1960
7 Armenia europe_central_asia 1960
8 Antigua and Barbuda america 1960
9 Australia east_asia_pacific 1960
10 Austria europe_central_asia 1960
# … with 9,833 more rows
In this case, the rows are sorted by increasing numeric order of the variable year.
When ordering by a character variable, the order will be determined alphabetically:
gapminder1960to2010 %>%
select(country, world_region, year) %>%
# order by world_region
arrange(world_region)
# A tibble: 9,843 x 3
country world_region year
<chr> <chr> <dbl>
1 Argentina america 1960
2 Argentina america 1961
3 Argentina america 1962
4 Argentina america 1963
5 Argentina america 1964
6 Argentina america 1965
7 Argentina america 1966
8 Argentina america 1967
9 Argentina america 1968
10 Argentina america 1969
# … with 9,833 more rows
To arrange based on descending order, you can wrap the variable in the desc()
function:
gapminder1960to2010 %>%
select(country, world_region, year) %>%
# order by year
arrange(desc(year))
# A tibble: 9,843 x 3
country world_region year
<chr> <chr> <dbl>
1 Afghanistan south_asia 2010
2 Angola sub_saharan_africa 2010
3 Albania europe_central_asia 2010
4 Andorra europe_central_asia 2010
5 United Arab Emirates middle_east_north_africa 2010
6 Argentina america 2010
7 Armenia europe_central_asia 2010
8 Antigua and Barbuda america 2010
9 Australia east_asia_pacific 2010
10 Austria europe_central_asia 2010
# … with 9,833 more rows
Finally, you can include several variables within arrange(), which will sort the
table by each of them. For example:
gapminder1960to2010 %>%
select(country, world_region, year) %>%
# order by year, then by world region (z-a), then country (z-a)
arrange(year, desc(world_region), desc(country))
# A tibble: 9,843 x 3
country world_region year
<chr> <chr> <dbl>
1 Zimbabwe sub_saharan_africa 1960
2 Zambia sub_saharan_africa 1960
3 Uganda sub_saharan_africa 1960
4 Togo sub_saharan_africa 1960
5 Tanzania sub_saharan_africa 1960
6 Sudan sub_saharan_africa 1960
7 South Sudan sub_saharan_africa 1960
8 South Africa sub_saharan_africa 1960
9 Somalia sub_saharan_africa 1960
10 Sierra Leone sub_saharan_africa 1960
# … with 9,833 more rows
Retain Unique Rows
Sometimes it is useful to retain rows with unique combinations of some of our variables
(i.e. remove any duplicated rows). This can be done with the distinct() function.
# get unique combination of main religion and world region
gapminder1960to2010 %>%
distinct(main_religion, world_region)
# A tibble: 27 x 2
world_region main_religion
<chr> <chr>
1 south_asia muslim
2 south_asia Muslim
3 sub_saharan_africa christian
4 sub_saharan_africa Christian
5 europe_central_asia muslim
6 europe_central_asia Muslim
7 europe_central_asia christian
8 europe_central_asia Christian
9 middle_east_north_africa muslim
10 middle_east_north_africa Muslim
# … with 17 more rows
Choosing Rows Based on Conditions
To choose rows based on specific criteria, we can use filter(). For example,
to get all the rows of data from the 2000’s:
gapminder1960to2010 %>%
select(country, year) %>%
filter(year > 2000)
# A tibble: 1,930 x 2
country year
<chr> <dbl>
1 Afghanistan 2001
2 Afghanistan 2002
3 Afghanistan 2003
4 Afghanistan 2004
5 Afghanistan 2005
6 Afghanistan 2006
7 Afghanistan 2007
8 Afghanistan 2008
9 Afghanistan 2009
10 Afghanistan 2010
# … with 1,920 more rows
Conditional Operations
It is important to understand that when we set a condition like above, the output is a logical vector. Let’s see an example using a small vector.
some_years <- c(1985, 1990, 1999, 1995, 2010, 2000)
some_years < 2000
[1] TRUE TRUE TRUE TRUE FALSE FALSE
So what the filter() function does is evaluate the condition and return the rows
for which that condition is true.
It is possible to combine several conditions by using the logical operators
& (AND) and | (OR). For example, if we wanted the years between 1990 and 2000:
# both conditions have to be true
some_years > 1990 & some_years < 2000
[1] FALSE FALSE TRUE TRUE FALSE FALSE
And if we wanted the years below 1990 or above 2000, then:
# only one or the other of the conditions has to be true
some_years < 1990 | some_years > 2000
[1] TRUE FALSE FALSE FALSE TRUE FALSE
Conditional operators
To set filtering conditions, use the following relational operators:
>is greater than>=is greater than or equal to<is less than<=is less than or equal to==is equal to!=is different from%in%is contained inTo combine conditions, use the following logical operators:
&AND|ORSome functions return logical results and can be used in filtering operations:
is.na(x)returns TRUE if a value in x is missingThe
!can be used to negate a logical condition:
!is.na(x)returns TRUE if a value in x is NOT missing!(x %in% y)returns TRUE if a value in x is NOT present in y
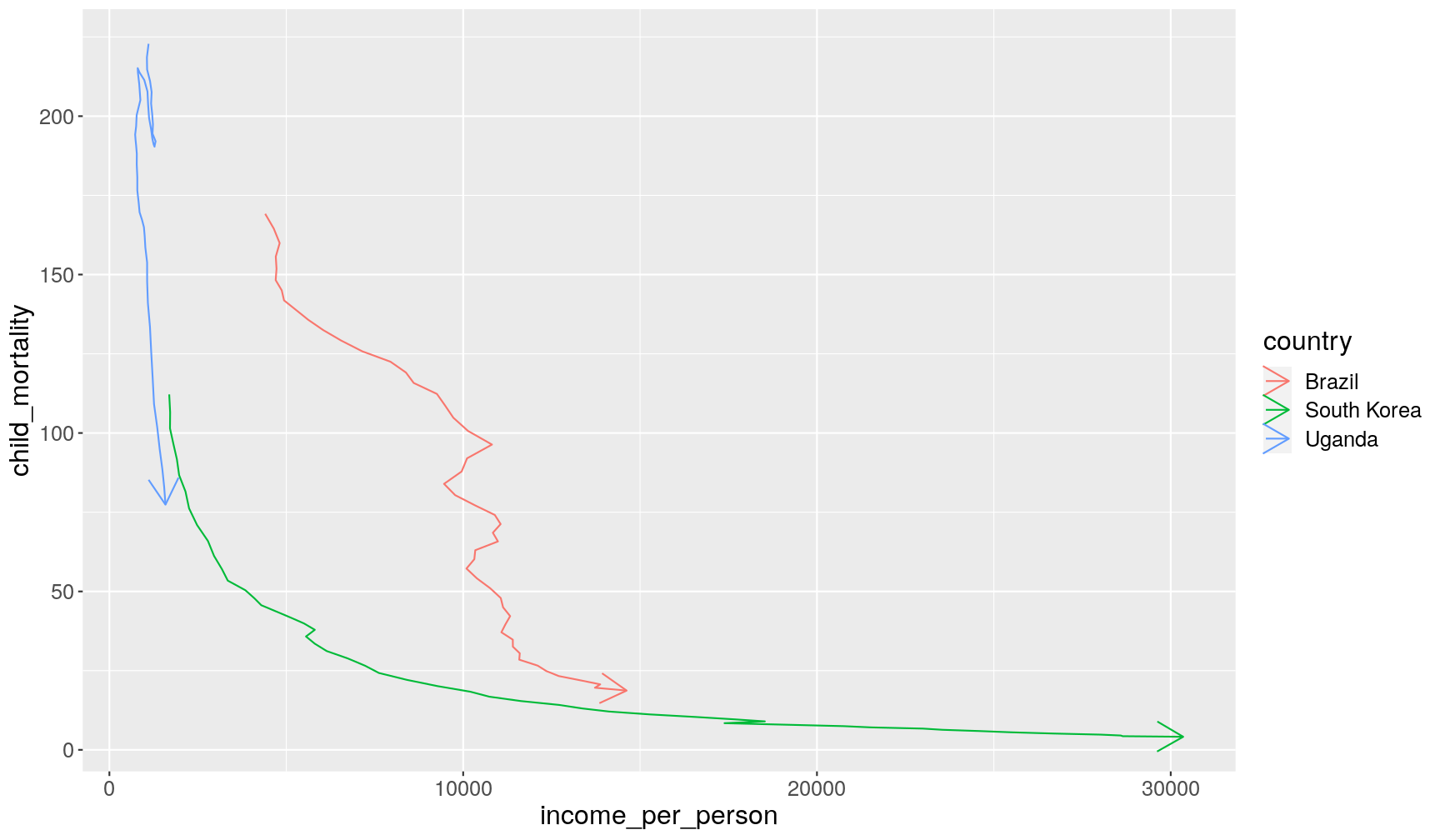
Let’s see an example on how we can combine the filter() function with plotting.
In Hans Rosling’s TED Talk,
they compared the change in income_per_person and child_mortality
between three countries: Uganda, Brazil and South Korea. We can filter our dataset
to retain only these countries by using the %in% operator.
gapminder1960to2010 %>%
filter(country %in% c("Uganda", "Brazil", "South Korea"))
We will pipe this filter to ggplot and use geom_path() to look at the change of these
countries over time.
This geometry is similar to geom_line(), but it connects points in the order they appear
on the table (which we can sort beforehand with arrange()), whereas geom_line() connects
the points based on the order they appear along the x and y axis.
(Try changing the code below to see the difference)
gapminder1960to2010 %>%
filter(country %in% c("Uganda", "Brazil", "South Korea")) %>%
# ensure data is in order of country and year for geom_path
arrange(country, year) %>%
ggplot(aes(income_per_person, child_mortality)) +
geom_path(aes(colour = country), arrow = arrow())
Exercise
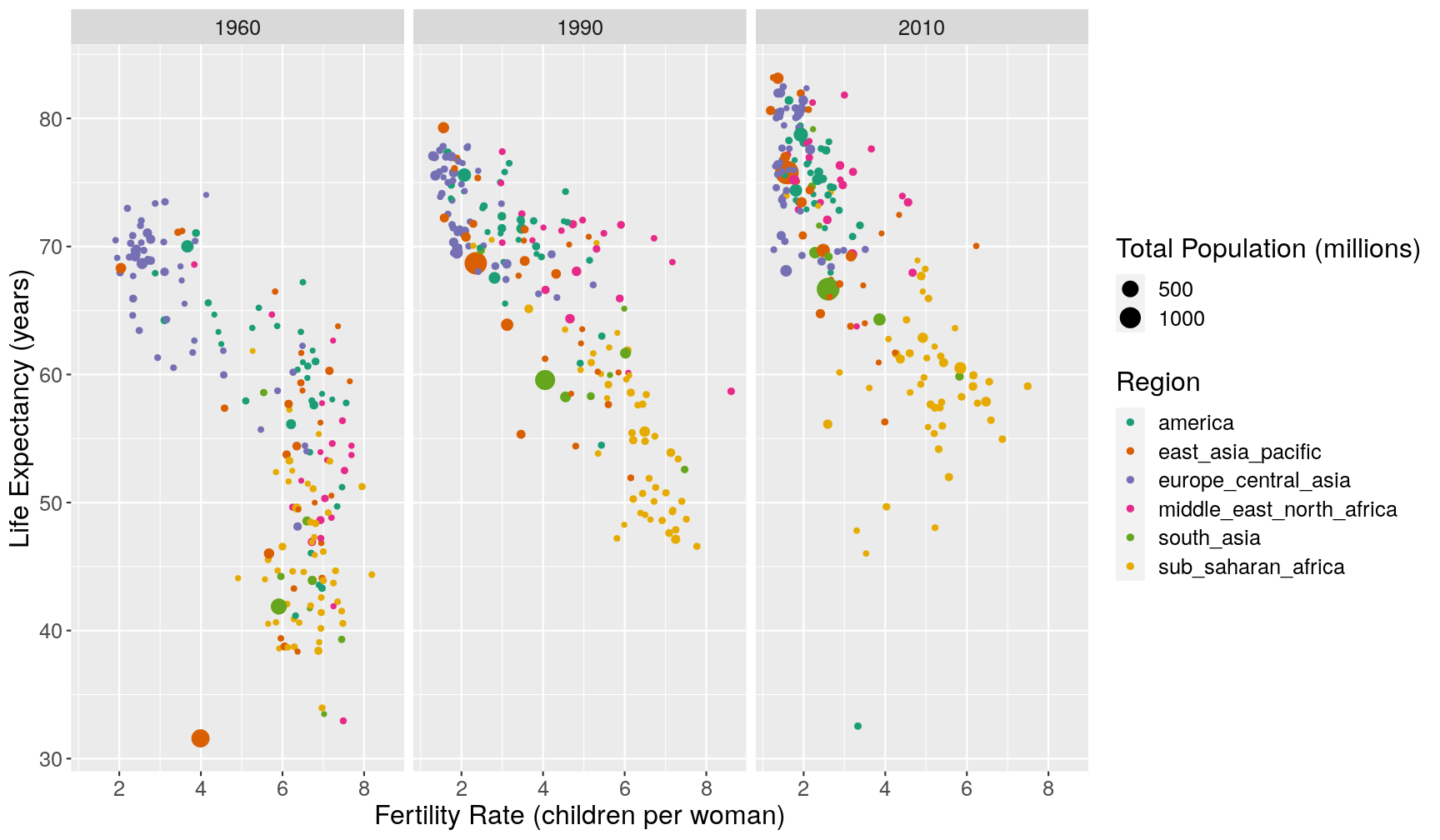
- Fix the following code (where the word “FIXME” appears), to produce the plot shown below.
gapminder1960to2010 %>% mutate(population_millions = FIXME) %>% filter(FIXME) %>% ggplot(aes(children_per_woman, life_expectancy)) + geom_point(aes(size = population_millions, colour = world_region)) + facet_grid(cols = FIXME) + scale_colour_brewer(palette = "Dark2") + labs(x = "Fertility Rate (children per woman)", y = "Life Expectancy (years)", size = "Total Population (millions)", colour = "Region")
- One of the countries seems to stand out from the general trend in 1960. Using some conditions inside
filter(), can you find what country it is?- Make a line plot showing how the
life_expectancyof that country changed over time compared to another country of your choice.Answer
A1. Here is the fixed code:
gapminder1960to2010 %>% mutate(population_millions = population/1e6) %>% filter(year %in% c(1960, 1990, 2010)) %>% ggplot(aes(children_per_woman, life_expectancy)) + geom_point(aes(size = population_millions, colour = world_region)) + facet_grid(cols = vars(year)) + scale_colour_brewer(palette = "Dark2") + labs(x = "Fertility Rate (children per woman)", y = "Life Expectancy (years)", size = "Total Population (millions)", colour = "Region")A2. Based on the x and y values in the graph, we can set a condition with
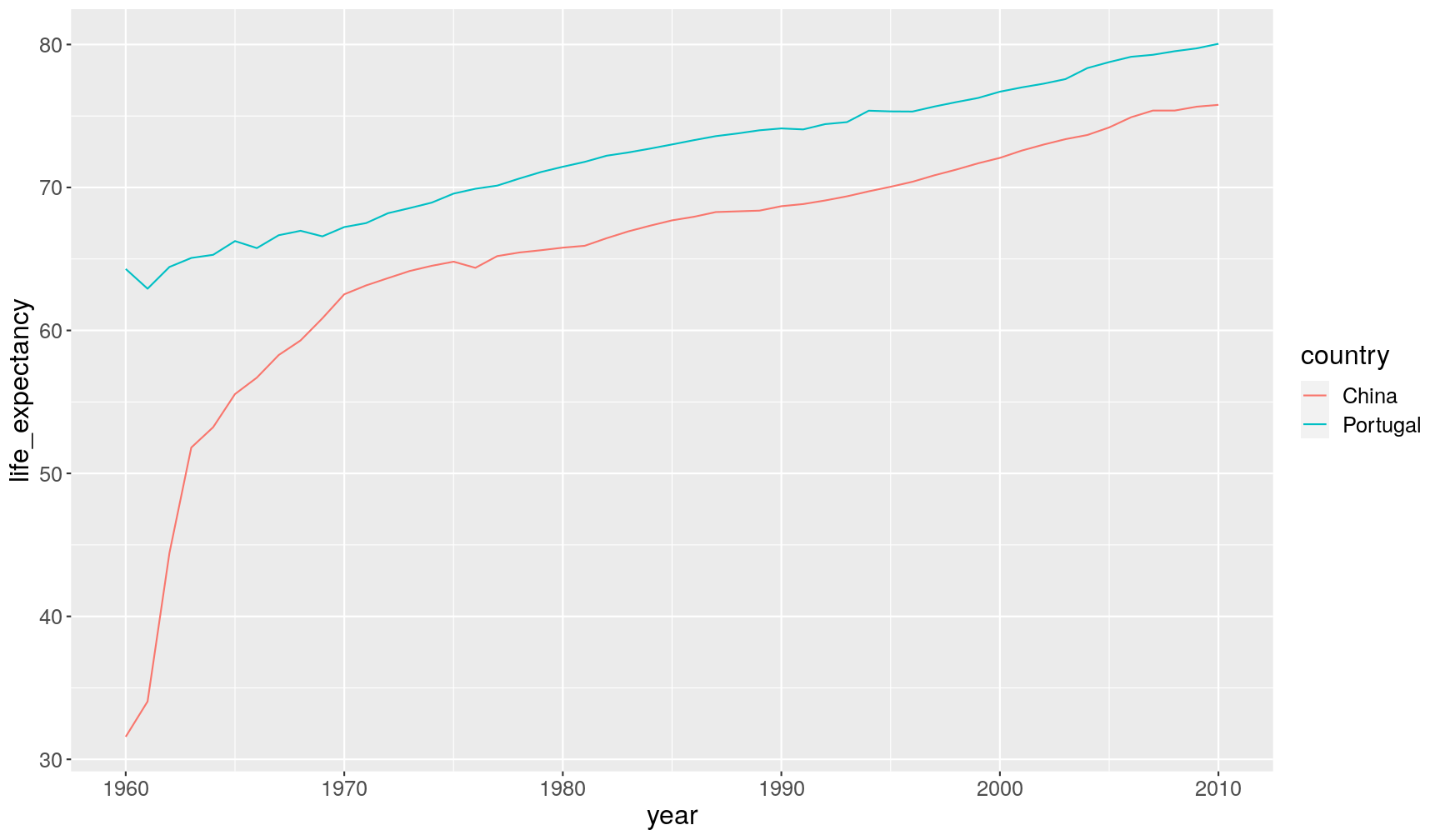
filter(), for example:gapminder1960to2010 %>% filter(year == 1960 & children_per_woman < 5 & life_expectancy < 35) %>% select(country, year, children_per_woman, life_expectancy)# A tibble: 1 x 4 country year children_per_woman life_expectancy <chr> <dbl> <dbl> <dbl> 1 China 1960 3.99 31.6A3. Here is a comparison between China and another country:
gapminder1960to2010 %>% filter(country %in% c("China", "Portugal")) %>% ggplot(aes(x = year, y = life_expectancy)) + geom_line(aes(colour = country))
Filtering Missing Values
As noted in the callout box above, the is.na() function can be used to ask the
question of whether a value is missing or not (!is.na()).
For example, the following would return the rows where main_religion is not
missing.
gapminder1960to2010 %>%
filter(!is.na(main_religion))
Exercise
- How many observations contain values for both
life_expectancyandchildren_per_woman?Answer
gapminder1960to2010 %>% filter(!is.na(life_expectancy) & !is.na(children_per_woman)) %>% # (optional) pipe the output to nrow() to get the number of rows directly nrow()[1] 9384
Using conditionals with ifelse
Conditions are not just useful for filtering rows, but also to modify variables or highlight certain observations in the data.
For example, take the following graph showing the change in income per person:
gapminder1960to2010 %>%
ggplot(aes(x = year, y = income_per_person)) +
geom_line(aes(group = country))
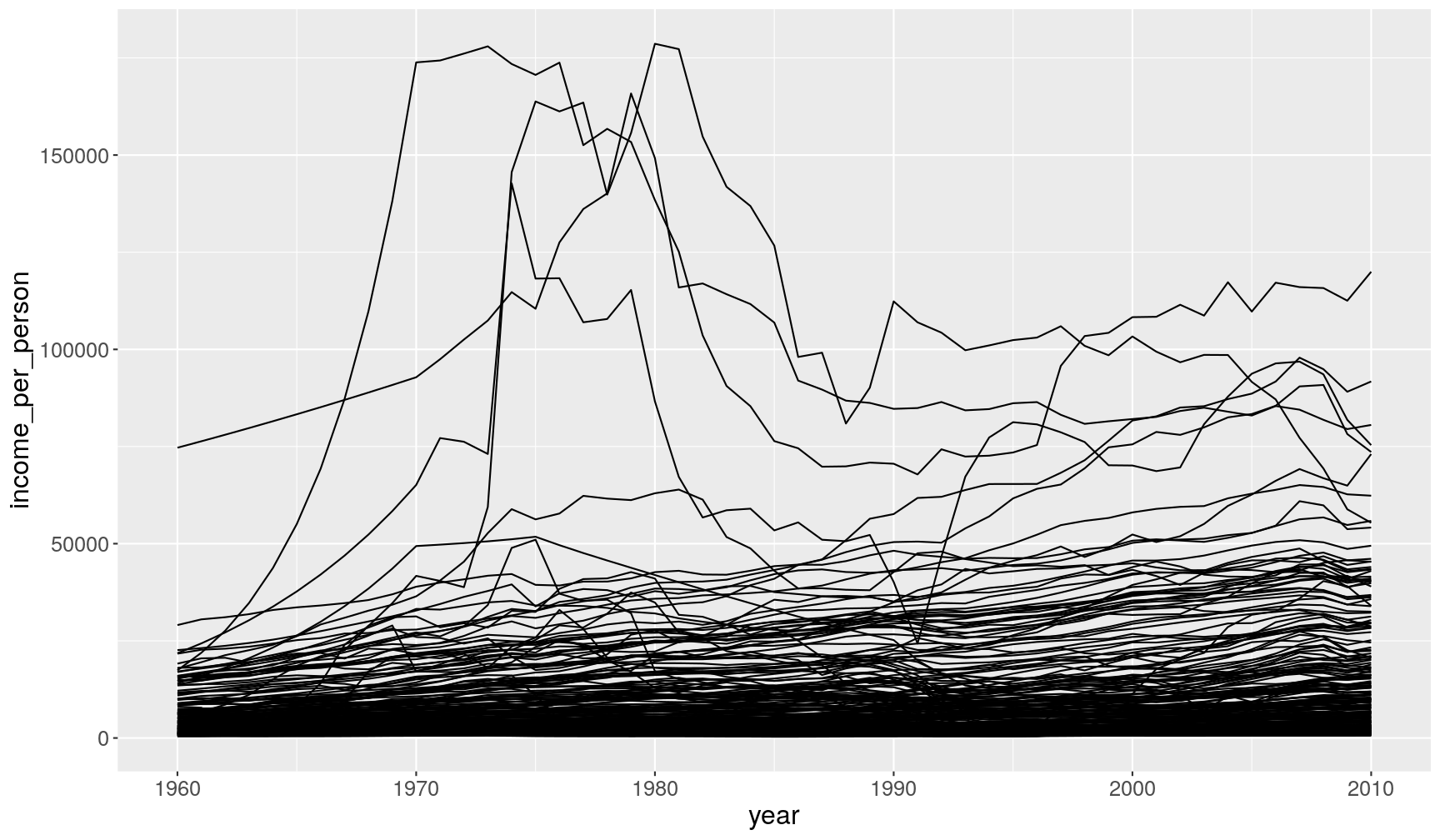
We can highlight one of the countries by creating a new column (mutate()) that stores
the result of a condition:
gapminder1960to2010 %>%
# the "is_qatar" column will contain TRUE/FALSE values
mutate(is_qatar = country == "Qatar") %>%
ggplot(aes(x = year, y = income_per_person)) +
geom_line(aes(group = country, colour = is_qatar))
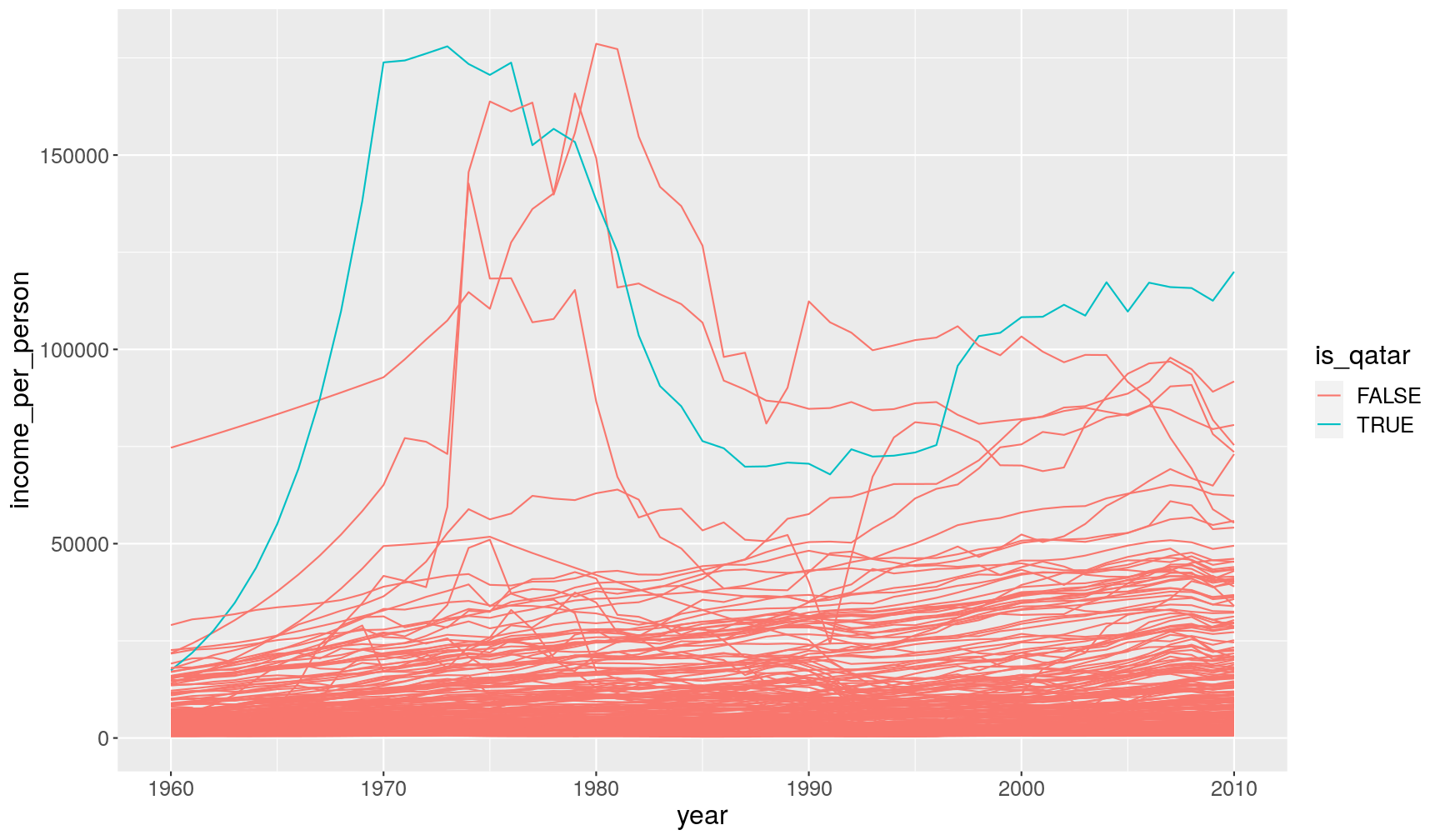
In this case, we used the logical output of that condition as the variable to colour our lines with.
However, sometimes we want more flexibility in defining outcomes from a condition,
which is where the ifelse() is handy.
This function takes 3 arguments: the condition we want to test,
the values it should take if it’s TRUE, and the values it should take if it’s FALSE.
For example, let’s highlight two countries in the graph and label all the remaining
countries as “Others” by using ifelse():
gapminder1960to2010 %>%
mutate(income_total = population*income_per_person,
country_highlight = ifelse(country %in% c("China", "United States"), country, "Others")) %>%
ggplot(aes(year, income_total)) +
geom_line(aes(group = country, colour = country_highlight))
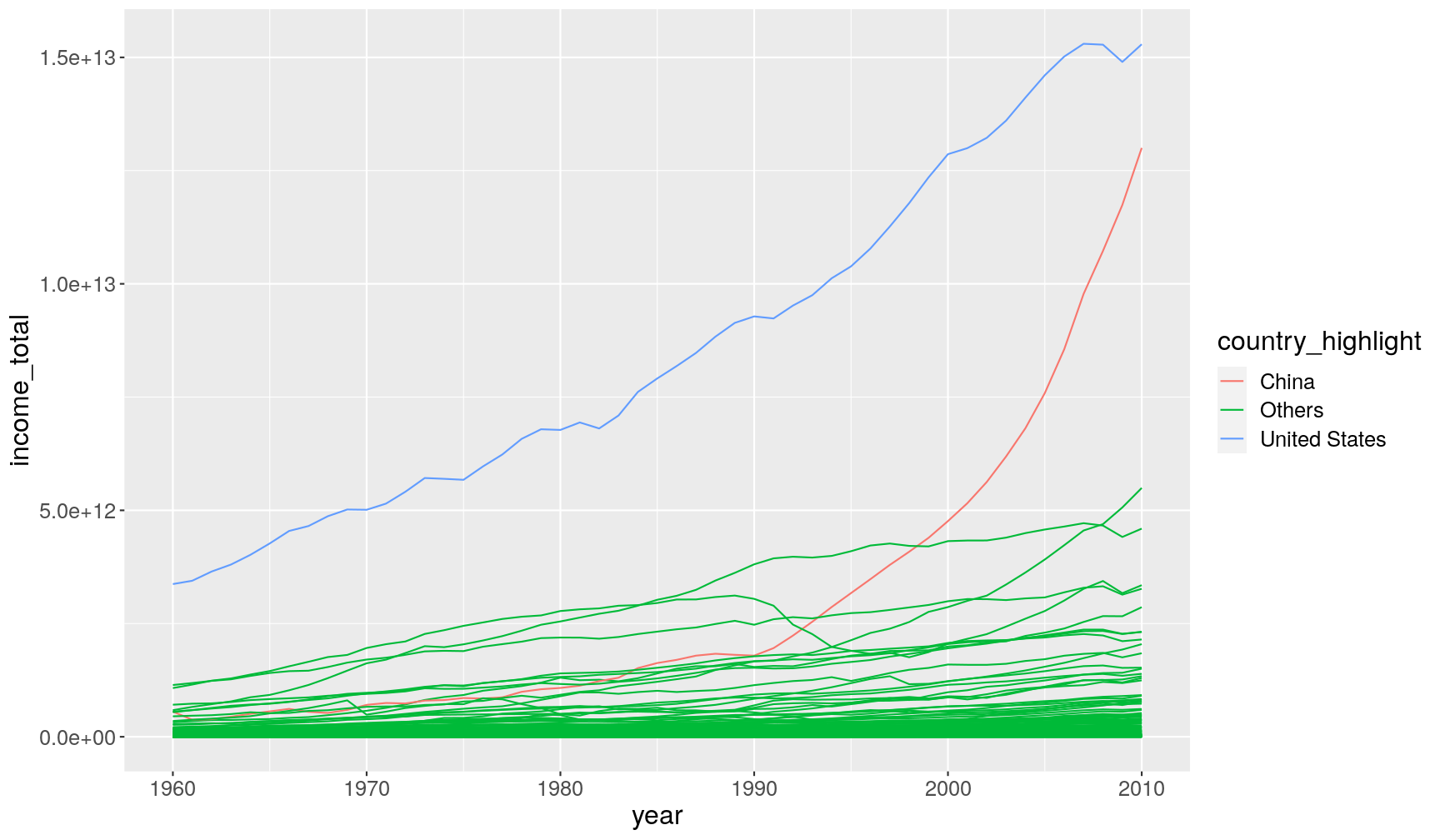
This is what ifelse() did for us:
if the value of country is either “China” or “United States” then return
their respective value, otherwise return the string “Others”.
ifelse() works in a vectorised manner, meaning that it evaluates the condition
at each value and returns the respective result we ask for.
Exercise
Earlier we saw that
life_expectancy_malehas some values encoded as “-999”:summary(gapminder1960to2010$life_expectancy_male)Min. 1st Qu. Median Mean 3rd Qu. Max. -999.00 50.03 61.73 10.64 68.15 80.08Try to fix this issue by using
ifelse()inside amutate(), replacing the values oflife_expectancy_malewithNAif they are equal to “-999” and keeping the original value otherwise.Save the result by updating
gapminder1960to2010and check that the issue is solved by using thesummary()function like above.Answer
# create new data.frame with missing values for life_expectancy_male gapminder1960to2010 <- gapminder1960to2010 %>% mutate(life_expectancy_male = ifelse(life_expectancy_male == -999, NA, life_expectancy_male)) # check the result summary(gapminder1960to2010$life_expectancy_male)Min. 1st Qu. Median Mean 3rd Qu. Max. NA's 16.29 51.80 62.56 60.02 68.41 80.08 459This is what
ifelse()did for us: if the value oflife_expectancy_maleis equal to -999 then return the valueNA, otherwise return the respective value oflife_expectancy_male.
Key Points
Order rows in a table using
arrange(). Use thedesc()function to sort in descending order.Retain unique rows in a table using
distinct().Choose rows based on conditions using
filter().Conditions can be set using several operators:
>,>=,<,<=,==,!=,%in%.Conditions can be combined using
&and|.The function
is.na()can be used to identify missing values. It can be negated as!is.na()to find non-missing values.Use the
ifelse()function to define two different outcomes of a condition.